Chopper Cascades#
Overview#
The chopper_cascade module provides utilities for computing time and wavelength bounds of a neutron pulse (or sub-pulses) propagating through a chopper cascade. This is useful for designing chopper systems, as well as predicting the data recorded when using techniques such as wavelength-frame multiplication (WFM).
It is currently under development and not fully functional.
Example: WFM chopper cascade#
As an example, consider the WFM chopper cascade from the tof package documentation.
[1]:
from scippneutron.tof import chopper_cascade
import scipp as sc
import matplotlib as mpl
mpl.rcParams['figure.dpi'] = 300
Defining choppers#
Choppers are defined by their position along the beam path, and the opening and closing times of their cutouts.
[2]:
params = {'wfm1': {'open': [-0.000396, 0.001286, 0.005786, 0.008039, 0.010133, 0.012080, 0.013889, 0.015571],
'close': [0.000654, 0.002464, 0.006222, 0.008646, 0.010899, 0.012993, 0.014939, 0.016750],
'distance': 6.6},
'wfm2': {'open': [0.000654, 0.002451, 0.006222, 0.008645, 0.010898, 0.012993, 0.014940, 0.016737],
'close': [0.001567, 0.003641, 0.006658, 0.009252, 0.011664, 0.013759, 0.015853, 0.017927],
'distance': 7.1},
'foc1': {'open': [-0.000139, 0.002460, 0.006796, 0.010020, 0.012733, 0.015263, 0.017718, 0.020317],
'close': [0.000640, 0.003671, 0.007817, 0.011171, 0.013814, 0.016146, 0.018497, 0.021528],
'distance': 8.8},
'foc2': {'open': [-0.000306, 0.010939, 0.016495, 0.021733, 0.026416, 0.030880, 0.035409],
'close': [0.002582, 0.014570, 0.020072, 0.024730, 0.029082, 0.033316, 0.038297],
'distance': 15.9}
}
choppers = sc.DataGroup(
{name: chopper_cascade.Chopper(
distance=sc.scalar(param['distance'], unit='m'),
time_open=sc.array(dims=('cutout',), values=param['open'], unit='s'),
time_close=sc.array(dims=('cutout',), values=param['close'], unit='s')
) for name, param in params.items()}
)
We can now initialize a frame-sequence and apply the chopper cascade to it:
[3]:
frames = chopper_cascade.FrameSequence.from_source_pulse(
time_min=sc.scalar(0.0, unit='ms'),
time_max=sc.scalar(4.0, unit='ms'), # ESS pulse is 3 ms, but it has a tail
wavelength_min=sc.scalar(0.0, unit='angstrom'),
wavelength_max=sc.scalar(10.0, unit='angstrom'),
)
frames = frames.chop(choppers.values())
at_sample = frames.propagate_to(sc.scalar(26.0, unit='m'))
at_sample.draw()
[3]:
(<Figure size 1920x1440 with 1 Axes>,
<Axes: title={'center': 'Frame propagation through chopper cascade'}, xlabel='ms', ylabel='Å'>)
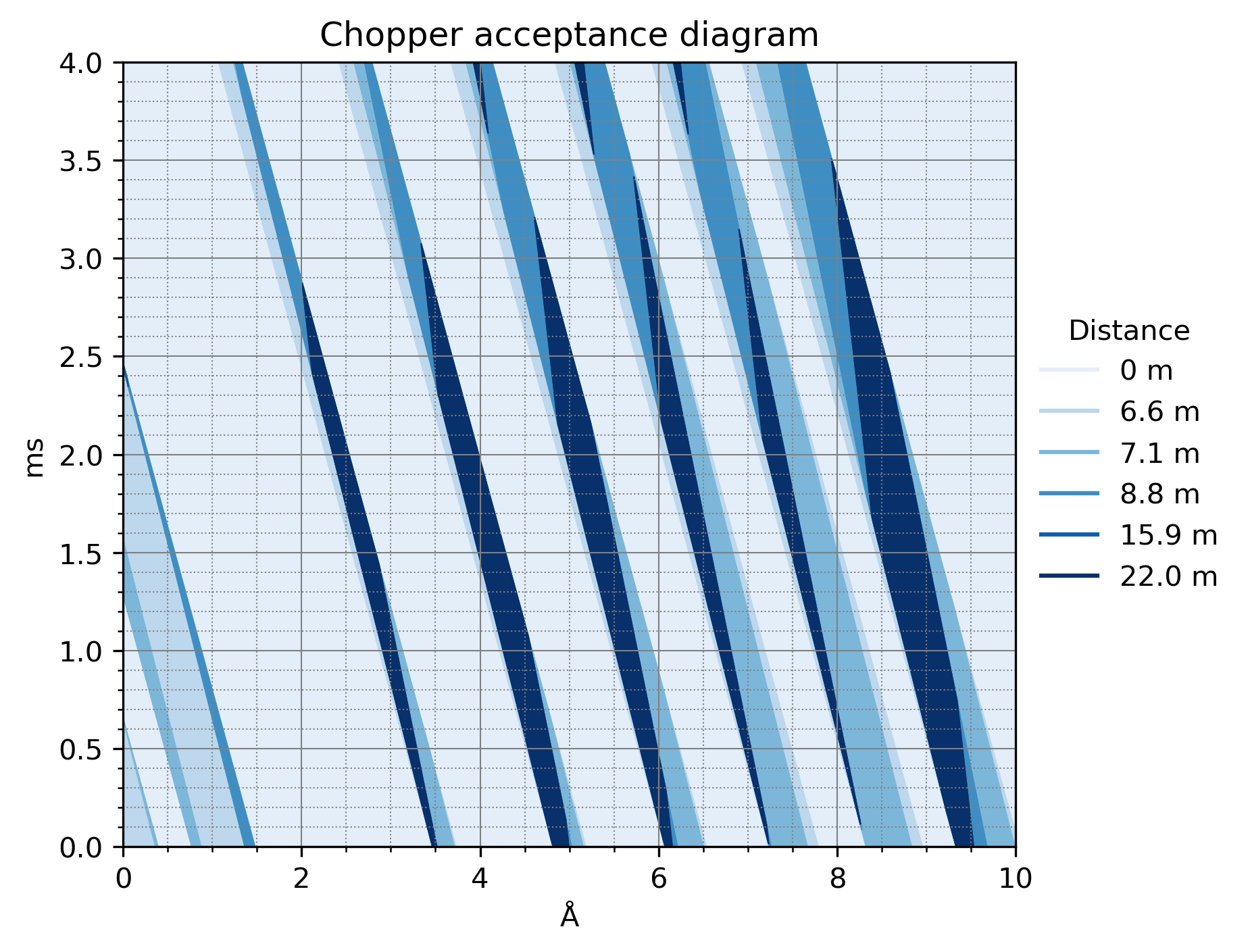
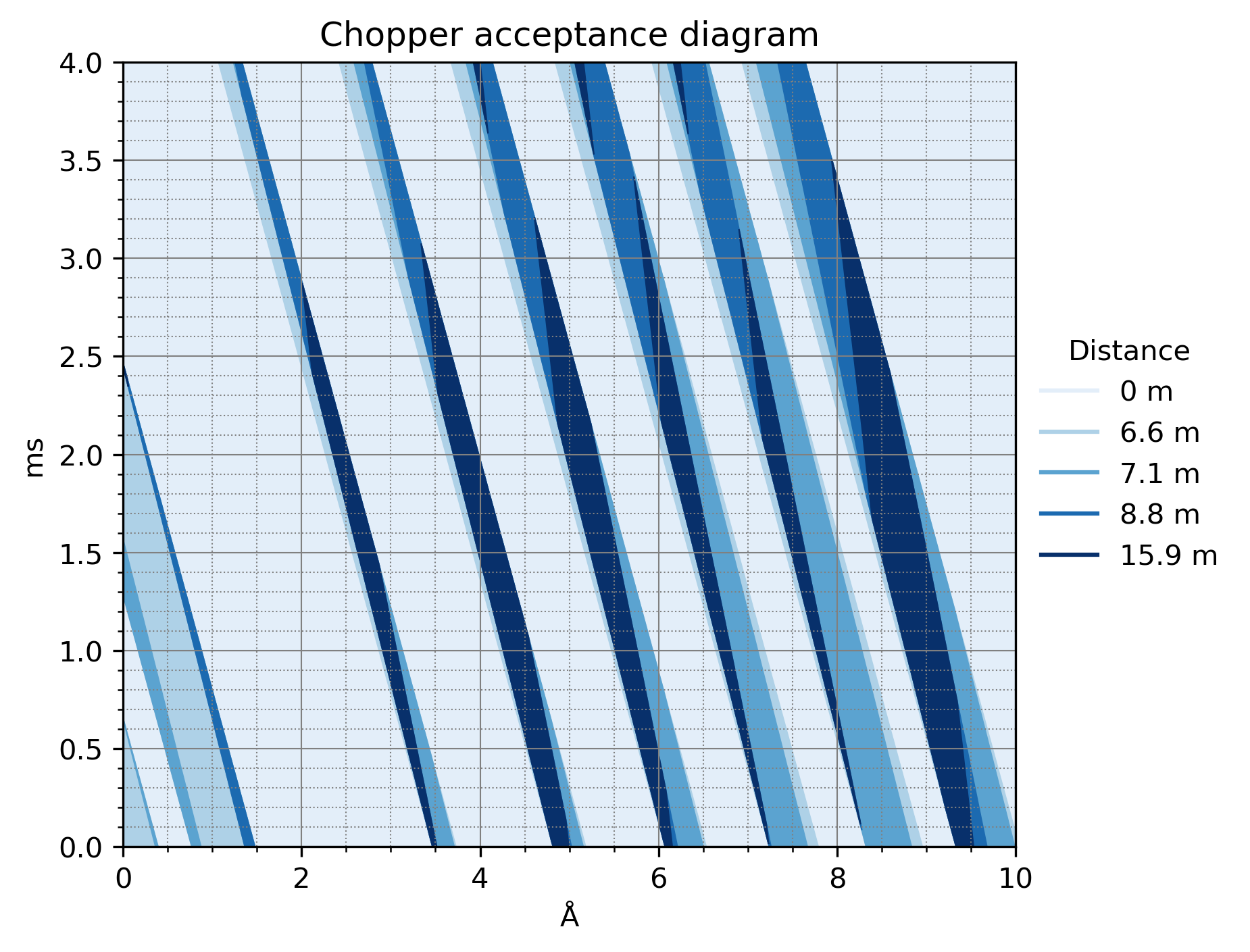
We can also draw a chopper acceptance diagram, which is essentially the same as above, but propagated back to the source pulse distance:
[4]:
frames.acceptance_diagram()
[4]:
(<Figure size 1920x1440 with 1 Axes>,
<Axes: title={'center': 'Chopper acceptance diagram'}, xlabel='Å', ylabel='ms'>)
Frame unwrapping#
For unwrapping frames, we need the bounds of the entire frame, to determine times at which to cut. Since \(L_2\) can be different for every detector, this cutting time is different for every detector. We can compute the frame bounds at a common distance, e.g., the sample, and propagate the bounds to the detectors:
[5]:
bounds = at_sample[-1].bounds()
chopper_cascade.propagate_times(
time=bounds['time'],
wavelength=bounds['wavelength'],
distance=sc.linspace('L2', 1.0, 2.0, 100, unit='m'),
)
[5]:
- (L2: 100, bound: 2)float64s0.002, 0.065, ..., 0.002, 0.067
Values:
array([[0.00246 , 0.06503264], [0.00246 , 0.06505697], [0.00246 , 0.0650813 ], [0.00246 , 0.06510563], [0.00246 , 0.06512996], [0.00246 , 0.06515429], [0.00246 , 0.06517862], [0.00246 , 0.06520295], [0.00246 , 0.06522728], [0.00246 , 0.06525161], [0.00246 , 0.06527594], [0.00246 , 0.06530027], [0.00246 , 0.0653246 ], [0.00246 , 0.06534892], [0.00246 , 0.06537325], [0.00246 , 0.06539758], [0.00246 , 0.06542191], [0.00246 , 0.06544624], [0.00246 , 0.06547057], [0.00246 , 0.0654949 ], [0.00246 , 0.06551923], [0.00246 , 0.06554356], [0.00246 , 0.06556789], [0.00246 , 0.06559222], [0.00246 , 0.06561655], [0.00246 , 0.06564088], [0.00246 , 0.06566521], [0.00246 , 0.06568954], [0.00246 , 0.06571387], [0.00246 , 0.0657382 ], [0.00246 , 0.06576253], [0.00246 , 0.06578685], [0.00246 , 0.06581118], [0.00246 , 0.06583551], [0.00246 , 0.06585984], [0.00246 , 0.06588417], [0.00246 , 0.0659085 ], [0.00246 , 0.06593283], [0.00246 , 0.06595716], [0.00246 , 0.06598149], [0.00246 , 0.06600582], [0.00246 , 0.06603015], [0.00246 , 0.06605448], [0.00246 , 0.06607881], [0.00246 , 0.06610314], [0.00246 , 0.06612747], [0.00246 , 0.0661518 ], [0.00246 , 0.06617613], [0.00246 , 0.06620046], [0.00246 , 0.06622478], [0.00246 , 0.06624911], [0.00246 , 0.06627344], [0.00246 , 0.06629777], [0.00246 , 0.0663221 ], [0.00246 , 0.06634643], [0.00246 , 0.06637076], [0.00246 , 0.06639509], [0.00246 , 0.06641942], [0.00246 , 0.06644375], [0.00246 , 0.06646808], [0.00246 , 0.06649241], [0.00246 , 0.06651674], [0.00246 , 0.06654107], [0.00246 , 0.0665654 ], [0.00246 , 0.06658973], [0.00246 , 0.06661406], [0.00246 , 0.06663839], [0.00246 , 0.06666272], [0.00246 , 0.06668704], [0.00246 , 0.06671137], [0.00246 , 0.0667357 ], [0.00246 , 0.06676003], [0.00246 , 0.06678436], [0.00246 , 0.06680869], [0.00246 , 0.06683302], [0.00246 , 0.06685735], [0.00246 , 0.06688168], [0.00246 , 0.06690601], [0.00246 , 0.06693034], [0.00246 , 0.06695467], [0.00246 , 0.066979 ], [0.00246 , 0.06700333], [0.00246 , 0.06702766], [0.00246 , 0.06705199], [0.00246 , 0.06707632], [0.00246 , 0.06710065], [0.00246 , 0.06712497], [0.00246 , 0.0671493 ], [0.00246 , 0.06717363], [0.00246 , 0.06719796], [0.00246 , 0.06722229], [0.00246 , 0.06724662], [0.00246 , 0.06727095], [0.00246 , 0.06729528], [0.00246 , 0.06731961], [0.00246 , 0.06734394], [0.00246 , 0.06736827], [0.00246 , 0.0673926 ], [0.00246 , 0.06741693], [0.00246 , 0.06744126]])
For WFM, we need to compute subframe time cutting points. Again, \(L_2\) can be different for every detector, so we need to compute the cutting points for every detector:
[6]:
bounds = at_sample[-1].subbounds()
bounds
[6]:
- timescippVariable(subframe: 10, bound: 2)float64s0.002, 0.003, ..., 0.056, 0.063
- wavelengthscippVariable(subframe: 10, bound: 2)float64Å0.0, 0.059, ..., 7.937, 9.529
[7]:
chopper_cascade.propagate_times(
time=bounds['time'],
wavelength=bounds['wavelength'],
distance=sc.linspace('L2', 1.0, 2.0, 100, unit='m'),
)
[7]:
- (L2: 100, subframe: 10, bound: 2)float64s0.002, 0.003, ..., 0.060, 0.067
Values:
array([[[0.00246 , 0.00274724], [0.01656897, 0.02398398], [0.02586316, 0.03408453], ..., [0.04608932, 0.04684825], [0.05023934, 0.05649482], [0.05767942, 0.06503264]], [[0.00246 , 0.00274739], [0.01657409, 0.02399295], [0.02587169, 0.03409728], ..., [0.04610506, 0.04686442], [0.05025696, 0.05651591], [0.05769969, 0.06505697]], [[0.00246 , 0.00274754], [0.01657921, 0.02400192], [0.02588021, 0.03411003], ..., [0.04612081, 0.04688059], [0.05027457, 0.056537 ], [0.05771995, 0.0650813 ]], ..., [[0.00246 , 0.00276182], [0.01706593, 0.02485433], [0.02669009, 0.03532142], ..., [0.04761669, 0.04841648], [0.05194819, 0.05854081], [0.05964523, 0.0673926 ]], [[0.00246 , 0.00276197], [0.01707105, 0.0248633 ], [0.02669861, 0.03533417], ..., [0.04763244, 0.04843265], [0.05196581, 0.05856191], [0.0596655 , 0.06741693]], [[0.00246 , 0.00276213], [0.01707617, 0.02487227], [0.02670714, 0.03534692], ..., [0.04764818, 0.04844882], [0.05198342, 0.058583 ], [0.05968576, 0.06744126]]])
Note that subframes may in principle overlap, and this may depend on the detector. We therefore call subframe_bounds at a common location and propagate the result, otherwise we would get a different number of subframes for different detectors.