Components#
The notebook will describe the different component that can be added to the beamline, their parameters, and how to inspect the neutrons that reach each component.
[1]:
import scipp as sc
import tof
meter = sc.Unit('m')
Hz = sc.Unit('Hz')
deg = sc.Unit('deg')
We begin by making a source pulse using the profile from ESS.
[2]:
source = tof.Source(facility='ess', neutrons=1_000_000)
source.plot()
Downloading file 'ess/ess.h5' from 'https://github.com/scipp/tof-sources/raw/refs/heads/main/1/ess/ess.h5' to '/home/runner/.cache/tof'.
[2]:
Adding a detector#
We first add a Detector component which simply records all the neutrons that reach it. It does not block any neutrons, they all travel through the detector without being absorbed.
[3]:
detector = tof.Detector(distance=30.0 * meter, name='detector')
# Build the instrument model
model = tof.Model(source=source, detectors=[detector])
model
[3]:
Model:
Source: Source:
pulses=1, neutrons per pulse=1000000
frequency=14.0 Hz
facility='ess'
distance=0.0 m
Choppers:
Detectors:
detector: Detector(name=detector, distance=30.0 m)
[4]:
# Run and plot the rays
res = model.run()
res.plot()
[4]:
Plot(ax=<Axes: xlabel='Time [μs]', ylabel='Distance [m]'>, fig=<Figure size 640x480 with 2 Axes>)
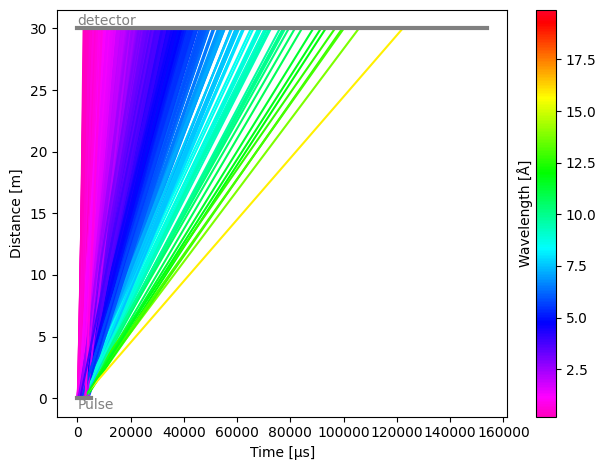
As expected, the detector sees all the neutrons from the pulse. Each component in the instrument has a .plot() method, which allows us to quickly visualize histograms of the neutron counts at the detector.
[5]:
res.detectors['detector'].plot()
[5]:
The data itself is available via the .toa, .wavelength, .birth_time, and .speed properties, depending on which one you wish to inspect.
[6]:
print(res.detectors['detector'].toa)
print(res.detectors['detector'].wavelength)
print(res.detectors['detector'].birth_time)
print(res.detectors['detector'].speed)
toa: min=1593.976873892107 µs, max=153524.63380991982 µs, events=1000000
wavelength: min=0.19851657277682427 Å, max=19.899992624660857 Å, events=1000000
birth_time: min=12.451499491324176 µs, max=4985.517769577258 µs, events=1000000
speed: min=198.79575237716256 m/s, max=19927.978560091717 m/s, events=1000000
Adding a chopper#
Next, we add a chopper in the beamline, with a frequency, phase, distance from source, and a set of open and close angles for the cutouts in the rotating disk.
[7]:
chopper1 = tof.Chopper(
frequency=10.0 * Hz,
open=sc.array(
dims=['cutout'],
values=[30.0, 50.0],
unit='deg',
),
close=sc.array(
dims=['cutout'],
values=[40.0, 80.0],
unit='deg',
),
phase=0.0 * deg,
distance=8 * meter,
name="Chopper1",
)
chopper1
[7]:
Chopper(name=Chopper1, distance=8.0 m, frequency=10.0 Hz, phase=0.0 deg, direction=CLOCKWISE, cutouts=2)
We can directly set this on our existing model, and re-run the simulation.
[8]:
model.add(chopper1)
res = model.run()
res.plot()
[8]:
Plot(ax=<Axes: xlabel='Time [μs]', ylabel='Distance [m]'>, fig=<Figure size 640x480 with 2 Axes>)
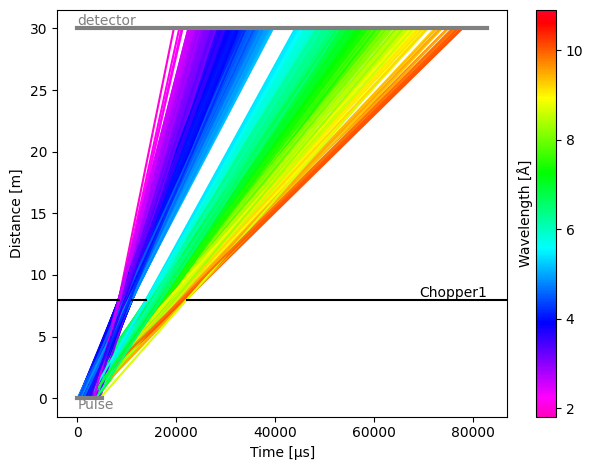
As expected, the two openings now create two bursts of neutrons, separating the wavelengths into two groups.
If we plot the chopper data,
[9]:
res.choppers['Chopper1'].toa.plot()
[9]:
we notice that the chopper sees all the incoming neutrons, and blocks many of them (gray), only allowing a subset to pass through the openings (blue).
The detector now sees two peaks in its histogrammed counts:
[10]:
res.detectors['detector'].toa.plot()
[10]:
Multiple choppers#
It is of course possible to add more than one chopper. Here we add a second one, further down the beam path, which splits each of the groups into two more groups.
[11]:
chopper2 = tof.Chopper(
frequency=5.0 * Hz,
open=sc.array(
dims=['cutout'],
values=[30.0, 40.0, 55.0, 70.0],
unit='deg',
),
close=sc.array(
dims=['cutout'],
values=[35.0, 48.0, 65.0, 90.0],
unit='deg',
),
phase=0.0 * deg,
distance=20 * meter,
name="Chopper2",
)
model.add(chopper2)
res = model.run()
res.plot()
[11]:
Plot(ax=<Axes: xlabel='Time [μs]', ylabel='Distance [m]'>, fig=<Figure size 640x480 with 2 Axes>)
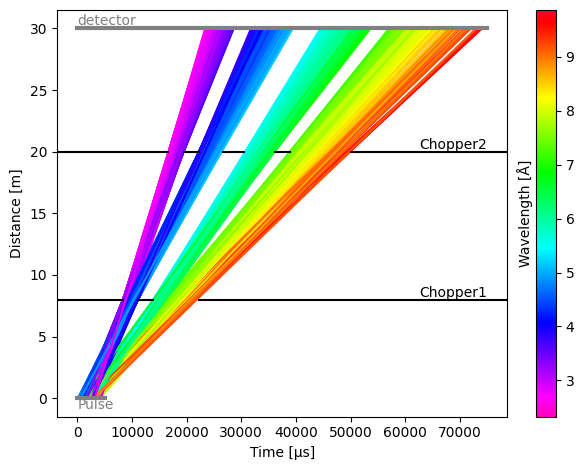
The distribution of neutrons that are blocked and pass through the second chopper looks as follows:
[12]:
res.choppers['Chopper2'].plot()
[12]:
while the detector now sees 4 peaks
[13]:
res.detectors['detector'].plot()
[13]:
To view the blocked rays on the time-distance diagram of the model, use
[14]:
res.plot(visible_rays=100, blocked_rays=5000)
[14]:
Plot(ax=<Axes: xlabel='Time [μs]', ylabel='Distance [m]'>, fig=<Figure size 640x480 with 2 Axes>)

Adding a monitor#
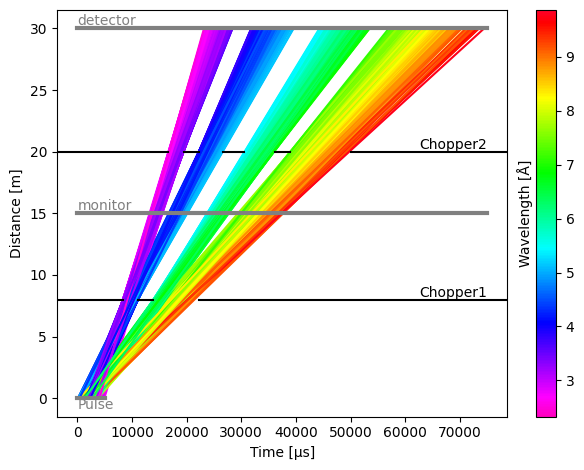
Detectors can be placed anywhere in the beam path, and in the next example we place a detector (which will act as a monitor) between the first and second chopper.
[15]:
monitor = tof.Detector(distance=15.0 * meter, name='monitor')
model.add(monitor)
res = model.run()
res.plot()
[15]:
Plot(ax=<Axes: xlabel='Time [μs]', ylabel='Distance [m]'>, fig=<Figure size 640x480 with 2 Axes>)
[16]:
res.detectors['monitor'].plot()
[16]:
Counter-rotating chopper#
By default, choppers are rotating clockwise. This means than when open and close angles of the chopper windows are defined as increasing angles in the anti-clockwise direction, the first window (with the lowest opening angles) will be the first one to pass in front of the beam.
To make a chopper rotate in the anti-clockwise direction, use the direction argument:
[17]:
chopper = tof.Chopper(
frequency=10.0 * Hz,
open=sc.array(
dims=['cutout'],
values=[280.0, 320.0],
unit='deg',
),
close=sc.array(
dims=['cutout'],
values=[310.0, 330.0],
unit='deg',
),
direction=tof.AntiClockwise,
phase=0.0 * deg,
distance=8 * meter,
name="Counter-rotating chopper",
)
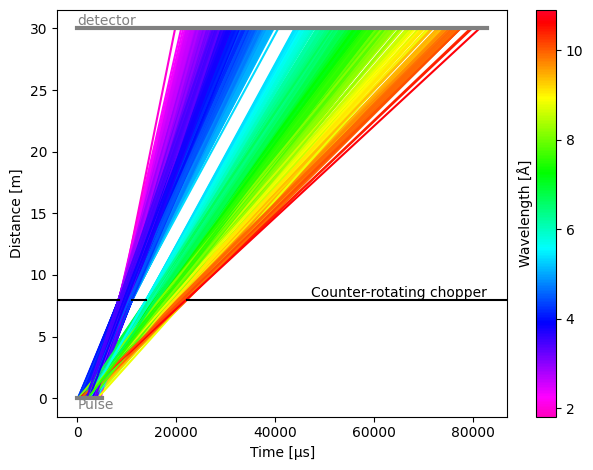
model = tof.Model(source=source, detectors=[detector], choppers=[chopper])
res = model.run()
[18]:
res.plot()
[18]:
Plot(ax=<Axes: xlabel='Time [μs]', ylabel='Distance [m]'>, fig=<Figure size 640x480 with 2 Axes>)
Loading from a JSON file#
It is also possible to load components from a JSON file, which can be very useful to quickly load a pre-configured instrument.
To illustrate the format, we first create a small JSON file and save it to disk:
[19]:
import json
params = {
"source": {
"type": "source",
"facility": "ess",
"neutrons": 1e6,
"pulses": 1
},
"chopper1": {
"type": "chopper",
"frequency": {"value": 56.0, "unit": "Hz"},
"phase": {"value": 93.244, "unit": "deg"},
"distance": {"value": 6.85, "unit": "m"},
"open": {
"value": [-1.9419, 49.5756, 98.9315, 146.2165, 191.5176, 234.9179],
"unit": "deg",
},
"close": {
"value": [1.9419, 55.7157, 107.2332, 156.5891, 203.8741, 249.1752],
"unit": "deg",
},
"direction": "clockwise",
},
"chopper2": {
"type": "chopper",
"frequency": {"value": 14.0, "unit": "Hz"},
"phase": {"value": 45.08, "unit": "deg"},
"distance": {"value": 8.45, "unit": "m"},
"open": {"value": [-23.6029], "unit": "deg"},
"close": {"value": [23.6029], "unit": "deg"},
"direction": "clockwise",
},
"detector": {"type": "detector", "distance": {"value": 60.5, "unit": "m"}},
}
with open("my_instrument.json", "w") as f:
json.dump(params, f)
[20]:
!head my_instrument.json
{"source": {"type": "source", "facility": "ess", "neutrons": 1000000.0, "pulses": 1}, "chopper1": {"type": "chopper", "frequency": {"value": 56.0, "unit": "Hz"}, "phase": {"value": 93.244, "unit": "deg"}, "distance": {"value": 6.85, "unit": "m"}, "open": {"value": [-1.9419, 49.5756, 98.9315, 146.2165, 191.5176, 234.9179], "unit": "deg"}, "close": {"value": [1.9419, 55.7157, 107.2332, 156.5891, 203.8741, 249.1752], "unit": "deg"}, "direction": "clockwise"}, "chopper2": {"type": "chopper", "frequency": {"value": 14.0, "unit": "Hz"}, "phase": {"value": 45.08, "unit": "deg"}, "distance": {"value": 8.45, "unit": "m"}, "open": {"value": [-23.6029], "unit": "deg"}, "close": {"value": [23.6029], "unit": "deg"}, "direction": "clockwise"}, "detector": {"type": "detector", "distance": {"value": 60.5, "unit": "m"}}}
We now use the tof.Model.from_json() method to load our instrument:
[21]:
model = tof.Model.from_json("my_instrument.json")
model
[21]:
Model:
Source: Source:
pulses=1, neutrons per pulse=1000000
frequency=14.0 Hz
facility='ess'
distance=0.0 m
Choppers:
chopper1: Chopper(name=chopper1, distance=6.85 m, frequency=56.0 Hz, phase=93.244 deg, direction=CLOCKWISE, cutouts=6)
chopper2: Chopper(name=chopper2, distance=8.45 m, frequency=14.0 Hz, phase=45.08 deg, direction=CLOCKWISE, cutouts=1)
Detectors:
detector: Detector(name=detector, distance=60.5 m)
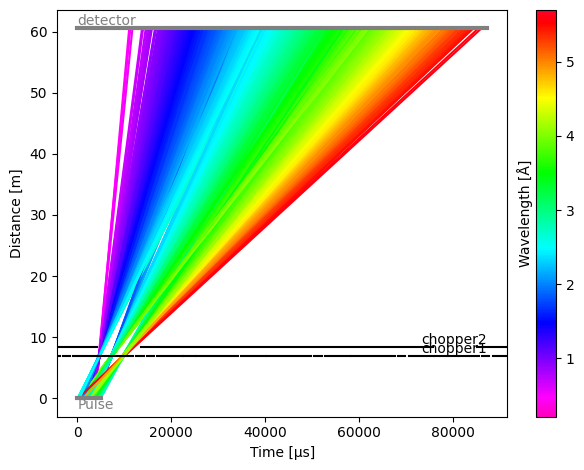
We can see that all components have been read in correctly. We can now run the model:
[22]:
model.run().plot()
[22]:
Plot(ax=<Axes: xlabel='Time [μs]', ylabel='Distance [m]'>, fig=<Figure size 640x480 with 2 Axes>)
Modifying the source#
The source in the instrument JSON file may need to be tweaked or swapped out entirely for a new one. Alternatively, the source may not even be specified in the JSON file.
To place a new source inside the model, simply create one and add it to the model:
[23]:
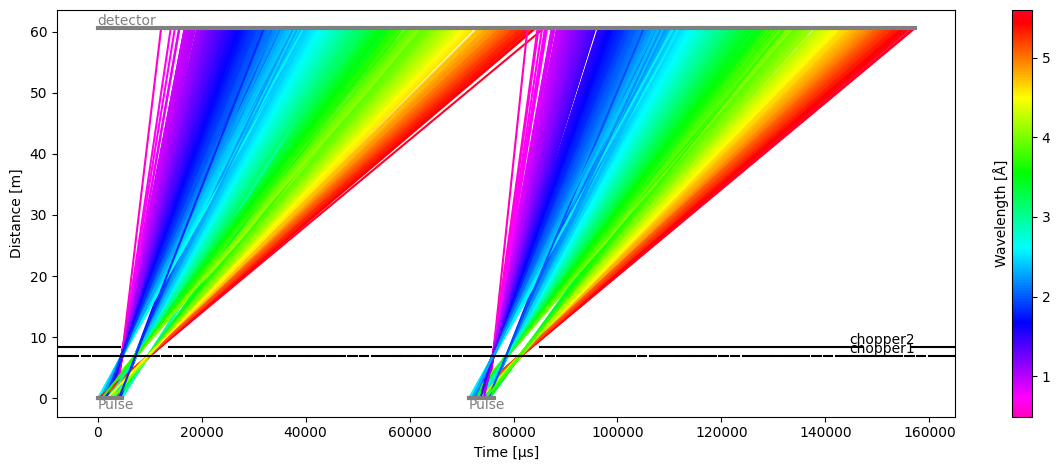
model.source = tof.Source(facility='ess', neutrons=1e4, pulses=2)
model.run().plot()
[23]:
Plot(ax=<Axes: xlabel='Time [μs]', ylabel='Distance [m]'>, fig=<Figure size 1200x480 with 2 Axes>)
Saving to JSON#
To share beamline parameters and models, you can save a model to a JSON file using the to_json method:
[24]:
model.to_json("new_instrument.json")