Bragg-edge imaging with ODIN#
This notebook illustrates how to convert recorded events on the ODIN detector to a single wavelength spectrum, revealing a Bragg edge in the data. WFM mode was used in the chopper cascade.
Loading dataset#
Loader is not part of
essimagingsince McStas dataset format is not stabilized yet.
[1]:
import scipp as sc
from ess.imaging.data import get_mcstas_ob_images_path, get_mcstas_sample_images_path
from odin_mcstas_helper import (
load_odin_simulation_data,
McStasManualResolution,
PLAIN_GRAPH,
)
from ess.reduce.nexus.types import FilePath
example_resolution = McStasManualResolution((128, 128))
# Small resolution for faster testing and documentation build.
ob_file_path = FilePath(get_mcstas_ob_images_path())
sample_file_path = FilePath(get_mcstas_sample_images_path())
ob_da = load_odin_simulation_data(ob_file_path, resolution=example_resolution)
sample_da = load_odin_simulation_data(sample_file_path, resolution=example_resolution)
sample_da
Downloading file 'README.md' from 'https://public.esss.dk/groups/scipp/ess/imaging/1/README.md' to '/home/runner/.cache/essimaging/1'.
Downloading file 'small_mcstas_ob_images.h5' from 'https://public.esss.dk/groups/scipp/ess/imaging/1/small_mcstas_ob_images.h5' to '/home/runner/.cache/essimaging/1'.
Downloading file 'small_mcstas_sample_images.h5' from 'https://public.esss.dk/groups/scipp/ess/imaging/1/small_mcstas_sample_images.h5' to '/home/runner/.cache/essimaging/1'.
[1]:
- event: 388566
- pixel_id(event)int64𝟙8122, 5408, ..., 15774, 969
Values:
array([ 8122, 5408, 4970, ..., 8714, 15774, 969], shape=(388566,)) - sample_position()vector3m[ 0. 0. 60.5]
Values:
array([ 0. , 0. , 60.5]) - sim_wavelength(event)float64Å5.727, 5.673, ..., 8.380, 6.448
Values:
array([5.72678488, 5.67272212, 3.17597509, ..., 7.567366 , 8.38023049, 6.44810452], shape=(388566,)) - source_position()vector3m[0. 0. 0.]
Values:
array([0., 0., 0.]) - time_of_arrival(event)float64µs8.994e+04, 8.894e+04, ..., 1.304e+05, 9.970e+04
Values:
array([ 89942.25084908, 88942.61380286, 50284.40890975, ..., 117180.5024966 , 130417.13999026, 99700.22439706], shape=(388566,))
- (event)float64counts22.133, 0.208, ..., 3.725, 0.688σ = 22.133, 0.208, ..., 3.725, 0.688
Values:
array([ 22.1326179 , 0.208402 , 198.90969649, ..., 0.56154171, 3.72506793, 0.68830282], shape=(388566,))
Variances (σ²):
array([4.89852775e+02, 4.34313955e-02, 3.95650674e+04, ..., 3.15329088e-01, 1.38761311e+01, 4.73760767e-01], shape=(388566,))
Convert McStas raw data to NeXus#
The raw McStas data looks different from what data in a NeXus file would look like. The time-of-flight recorded by the McStas monitor is a unwrapped time of arrival (see essreduce WFM workflow and scippneutron chopper cascade for more information); the tof coordinate has values beyond 71ms, as can be seen in the plot above.
The workflow that computes wavelengths from the WFM chopper cascade expects data in the NeXus format, so we transform the data here.
[2]:
from odin_mcstas_helper import to_nexus
sample_nexus = to_nexus(sample_da, resolution=example_resolution)
ob_nexus = to_nexus(ob_da, resolution=example_resolution)
sample_nexus
[2]:
- pixel_id: 16384
- Ltotal(pixel_id)float64m60.500, 60.500, ..., 60.500, 60.500
Values:
array([60.50001464, 60.50001442, 60.50001419, ..., 60.50001419, 60.50001442, 60.50001464], shape=(16384,)) - pixel_id(pixel_id)int64𝟙0, 1, ..., 16382, 16383
Values:
array([ 0, 1, 2, ..., 16381, 16382, 16383], shape=(16384,)) - position(pixel_id)vector3m[-2.9765625e-02 -2.9765625e-02 6.0500000e+01], [-2.9296875e-02 -2.9765625e-02 6.0500000e+01], ..., [2.9296875e-02 2.9765625e-02 6.0500000e+01], [2.9765625e-02 2.9765625e-02 6.0500000e+01]
Values:
array([[-2.9765625e-02, -2.9765625e-02, 6.0500000e+01], [-2.9296875e-02, -2.9765625e-02, 6.0500000e+01], [-2.8828125e-02, -2.9765625e-02, 6.0500000e+01], ..., [ 2.8828125e-02, 2.9765625e-02, 6.0500000e+01], [ 2.9296875e-02, 2.9765625e-02, 6.0500000e+01], [ 2.9765625e-02, 2.9765625e-02, 6.0500000e+01]], shape=(16384, 3)) - sample_position()vector3m[ 0. 0. 60.5]
Values:
array([ 0. , 0. , 60.5]) - source_position()vector3m[0. 0. 0.]
Values:
array([0., 0., 0.])
- (pixel_id)float64countsbinned data [len=36, len=29, ..., len=40, len=37]
dim='event', content=DataArray( dims=(event: 388566), data=float64[counts], coords={'sim_wavelength':float64[Å], 'event_time_zero':datetime64[ns], 'event_time_offset':float64[µs]})
[3]:
# Visualize
fig_sample = (
sample_nexus.bins.concat()
.hist(event_time_offset=300)
.plot(title='McStas simulation: sample\n(wrapped like ESS format, 14Hz pulse)')
)
fig_ob = (
ob_nexus.bins.concat()
.hist(event_time_offset=300)
.plot(title='McStas simulation: open beam\n(wrapped like ESS format, 14Hz pulse)')
)
fig_sample + fig_ob
[3]:
[4]:
import plopp as pp
pp.scatter3d(sample_nexus.hist(), pos='position', cbar=True, pixel_size=0.0005)
[4]:
Choppers#
To accurately compute the wavelengths of the neutrons from their time-of-arrival, we need the parameters of the choppers in the beamline.
[5]:
import sciline as sl
from odin_mcstas_helper import DISK_CHOPPERS
disk_choppers = DISK_CHOPPERS.copy() # Copy to avoid modifying the original dictionary
[6]:
disk_choppers["WFMC_1"]
[6]:
- axle_positionscippVariable()vector3m[0. 0. 6.85]
- frequencyscippVariable()float64Hz-56.0
- beam_positionscippVariable()float64deg0.0
- phasescippVariable()float64deg-93.244
- slit_beginscippVariable(cutout: 6)float64deg-1.942, 49.576, ..., 191.518, 234.918
- slit_endscippVariable(cutout: 6)float64deg1.942, 55.716, ..., 203.874, 249.175
- slit_heightNoneType()None
- radiusNoneType()None
Check that the chopper settings make sense with a quick tof run#
As useful sanity check is to run a basic simulation, propagating neutrons through the chopper cascade, using the Tof package.
[7]:
from ess.reduce.time_of_flight.fakes import FakeBeamline
Ltotal = sample_nexus.coords['Ltotal'].mean()
ess_beamline = FakeBeamline(
choppers=disk_choppers,
monitors={"detector": Ltotal},
run_length=sc.scalar(1 / 14, unit="s") * 8,
events_per_pulse=100_000,
)
ess_beamline.model_result.plot()
[7]:
Plot(ax=<Axes: xlabel='Time [μs]', ylabel='Distance [m]'>, fig=<Figure size 1200x480 with 2 Axes>)
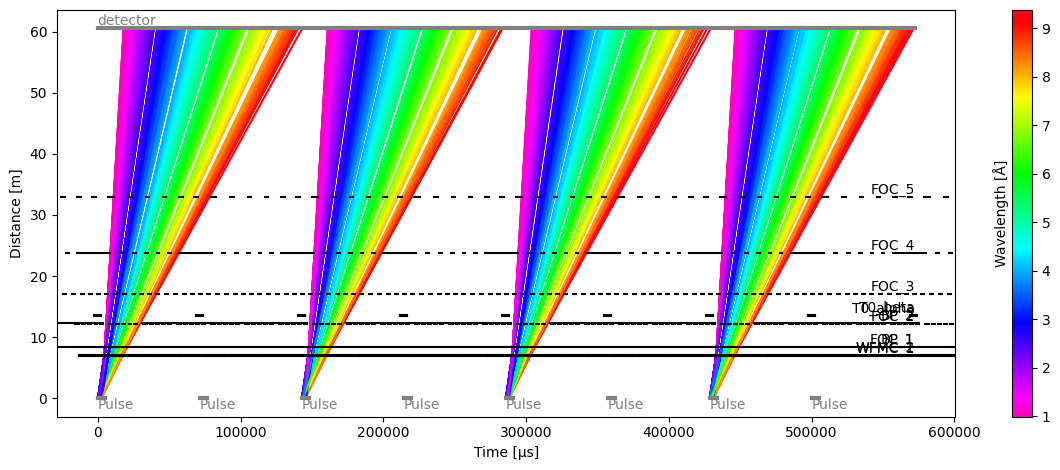
We observe that the WFM choppers make 6 distinct frames at the detector, and that the other choppers skip every other pulse to maximize wavelength coverage.
We can now compare the counts on the detector to our raw data, to make sure they broadly resemble each other.
[8]:
raw_data = ess_beamline.get_monitor("detector")[0]
# Visualize
fig_ob + raw_data.hist(event_time_offset=300).squeeze().plot(
title='Tof simulation (no sample)'
)
[8]:
Use WFM workflow#
We now set up the workflow which will convert the raw neutron arrival times to a real time-of-flight, and thus a wavelength.
Setting up the workflow#
[9]:
from ess.reduce import time_of_flight
workflow = sl.Pipeline(
time_of_flight.providers(), params=time_of_flight.default_parameters()
)
workflow[time_of_flight.RawData] = sample_nexus
workflow[time_of_flight.Ltotal] = sample_nexus.coords['Ltotal']
workflow[time_of_flight.LtotalRange] = (
sc.scalar(55.0, unit="m"),
sc.scalar(65.0, unit="m"),
)
workflow[time_of_flight.PulseStride] = 2 # Need for pulse-skipping
workflow[time_of_flight.SimulationResults] = time_of_flight.simulate_beamline(
choppers=disk_choppers, neutrons=2_000_000, source_position=sc.vector([0, 0, 0], unit='m')
)
workflow.visualize(time_of_flight.TofData)
[9]:
Inspect the lookup table#
[10]:
table = workflow.compute(time_of_flight.TimeOfFlightLookupTable).squeeze()
table['pulse', 0].plot(title='Pulse 0') + table['pulse', 1].plot(title='Pulse 1')
[10]:
Compute neutron time-of-flight#
[11]:
sample_tofs = workflow.compute(time_of_flight.TofData)
sample_tofs
[11]:
- pixel_id: 16384
- Ltotal(pixel_id)float64m60.500, 60.500, ..., 60.500, 60.500
Values:
array([60.50001464, 60.50001442, 60.50001419, ..., 60.50001419, 60.50001442, 60.50001464], shape=(16384,)) - pixel_id(pixel_id)int64𝟙0, 1, ..., 16382, 16383
Values:
array([ 0, 1, 2, ..., 16381, 16382, 16383], shape=(16384,)) - position(pixel_id)vector3m[-2.9765625e-02 -2.9765625e-02 6.0500000e+01], [-2.9296875e-02 -2.9765625e-02 6.0500000e+01], ..., [2.9296875e-02 2.9765625e-02 6.0500000e+01], [2.9765625e-02 2.9765625e-02 6.0500000e+01]
Values:
array([[-2.9765625e-02, -2.9765625e-02, 6.0500000e+01], [-2.9296875e-02, -2.9765625e-02, 6.0500000e+01], [-2.8828125e-02, -2.9765625e-02, 6.0500000e+01], ..., [ 2.8828125e-02, 2.9765625e-02, 6.0500000e+01], [ 2.9296875e-02, 2.9765625e-02, 6.0500000e+01], [ 2.9765625e-02, 2.9765625e-02, 6.0500000e+01]], shape=(16384, 3)) - sample_position()vector3m[ 0. 0. 60.5]
Values:
array([ 0. , 0. , 60.5]) - source_position()vector3m[0. 0. 0.]
Values:
array([0., 0., 0.])
- (pixel_id)float64countsbinned data [len=36, len=29, ..., len=40, len=37]
dim='event', content=DataArray( dims=(event: 388566), data=float64[counts], coords={'sim_wavelength':float64[Å], 'event_time_zero':datetime64[ns], 'event_time_offset':float64[µs], 'tof':float64[µs]})
[12]:
sample_wavs = sample_tofs.transform_coords('wavelength', graph=PLAIN_GRAPH)
sample_wavs
[12]:
- pixel_id: 16384
- Ltotal(pixel_id)float64m60.500, 60.500, ..., 60.500, 60.500
Values:
array([60.50001464, 60.50001442, 60.50001419, ..., 60.50001419, 60.50001442, 60.50001464], shape=(16384,)) - pixel_id(pixel_id)int64𝟙0, 1, ..., 16382, 16383
Values:
array([ 0, 1, 2, ..., 16381, 16382, 16383], shape=(16384,)) - position(pixel_id)vector3m[-2.9765625e-02 -2.9765625e-02 6.0500000e+01], [-2.9296875e-02 -2.9765625e-02 6.0500000e+01], ..., [2.9296875e-02 2.9765625e-02 6.0500000e+01], [2.9765625e-02 2.9765625e-02 6.0500000e+01]
Values:
array([[-2.9765625e-02, -2.9765625e-02, 6.0500000e+01], [-2.9296875e-02, -2.9765625e-02, 6.0500000e+01], [-2.8828125e-02, -2.9765625e-02, 6.0500000e+01], ..., [ 2.8828125e-02, 2.9765625e-02, 6.0500000e+01], [ 2.9296875e-02, 2.9765625e-02, 6.0500000e+01], [ 2.9765625e-02, 2.9765625e-02, 6.0500000e+01]], shape=(16384, 3)) - sample_position()vector3m[ 0. 0. 60.5]
Values:
array([ 0. , 0. , 60.5]) - source_position()vector3m[0. 0. 0.]
Values:
array([0., 0., 0.])
- (pixel_id)float64countsbinned data [len=36, len=29, ..., len=40, len=37]
dim='event', content=DataArray( dims=(event: 388566), data=float64[counts], coords={'sim_wavelength':float64[Å], 'event_time_zero':datetime64[ns], 'event_time_offset':float64[µs], 'tof':float64[µs], 'wavelength':float64[Å]})
We can now compare our computed wavelengths to the true wavelengths of the neutrons in the McStas simulation:
[13]:
true_wavs = sample_da.hist(sim_wavelength=300).rename(sim_wavelength='wavelength')
pp.plot(
{
'true': true_wavs,
'wfm': sample_wavs.bins.concat().hist(
wavelength=true_wavs.coords['wavelength']
),
},
title="ODIN McStas simulation",
)
[13]:
Region of interest#
Looking at the counts on the 2d detector panel, we see that there is a central rectangular darker region, surrounded by brighter edges.
[14]:
sample_folded = sample_wavs.fold(dim='pixel_id', sizes={'y': 128, 'x': 128})
sample_folded.hist().plot(aspect='equal')
[14]:
The dark region is where the beam was absorbed by the sample, and this is the region of interest. The brighter edges need to be discarded.
We crop the data using simple array slicing:
[15]:
sel = slice(11, 116, 1)
sample_cropped = sample_folded['y', sel]['x', sel]
sample_cropped.hist().plot(aspect='equal')
[15]:
Repeat for the open-beam#
We repeat the conversion to wavelength and crop the edges of the open-beam measurement.
[16]:
# Give the same pixel positions to both sample and open beam.
# Note: this is only because of the way we computed the positions.
# In practice, the geometry should come from the nexus file and this won't be needed.
ob_nexus.coords.update(
{key: sample_nexus.coords[key] for key in ('position', 'Ltotal')}
)
workflow[time_of_flight.Ltotal] = ob_nexus.coords['Ltotal']
workflow[time_of_flight.RawData] = ob_nexus
ob_tofs = workflow.compute(time_of_flight.TofData)
ob_wavs = ob_tofs.transform_coords('wavelength', graph=PLAIN_GRAPH)
ob_folded = ob_wavs.fold(dim='pixel_id', sizes={'y': 128, 'x': 128})
ob_cropped = ob_folded['y', sel]['x', sel]
Normalize the signal#
Finally, we are able to normalize our sample measurement to the open-beam data.
Here, we sum over all pixels before normalizing. There is no spatial structure in the signal, and we are only interested in the wavelength spectrum (where the Bragg edge is). So this is effectively like degrading the detector resolution to a single pixel.
[17]:
# Common set of bins
bins = sc.linspace('wavelength', 1.1, 9.5, 301, unit='angstrom')
num = sample_cropped.bins.concat().hist(wavelength=bins)
den = ob_cropped.bins.concat().hist(wavelength=bins)
normalized = num / den
normalized
[17]:
- wavelength: 300
- sample_position()vector3m[ 0. 0. 60.5]
Values:
array([ 0. , 0. , 60.5]) - source_position()vector3m[0. 0. 0.]
Values:
array([0., 0., 0.]) - wavelength(wavelength [bin-edge])float64Å1.1, 1.128, ..., 9.472, 9.5
Values:
array([1.1 , 1.128, 1.156, 1.184, 1.212, 1.24 , 1.268, 1.296, 1.324, 1.352, 1.38 , 1.408, 1.436, 1.464, 1.492, 1.52 , 1.548, 1.576, 1.604, 1.632, 1.66 , 1.688, 1.716, 1.744, 1.772, 1.8 , 1.828, 1.856, 1.884, 1.912, 1.94 , 1.968, 1.996, 2.024, 2.052, 2.08 , 2.108, 2.136, 2.164, 2.192, 2.22 , 2.248, 2.276, 2.304, 2.332, 2.36 , 2.388, 2.416, 2.444, 2.472, 2.5 , 2.528, 2.556, 2.584, 2.612, 2.64 , 2.668, 2.696, 2.724, 2.752, 2.78 , 2.808, 2.836, 2.864, 2.892, 2.92 , 2.948, 2.976, 3.004, 3.032, 3.06 , 3.088, 3.116, 3.144, 3.172, 3.2 , 3.228, 3.256, 3.284, 3.312, 3.34 , 3.368, 3.396, 3.424, 3.452, 3.48 , 3.508, 3.536, 3.564, 3.592, 3.62 , 3.648, 3.676, 3.704, 3.732, 3.76 , 3.788, 3.816, 3.844, 3.872, 3.9 , 3.928, 3.956, 3.984, 4.012, 4.04 , 4.068, 4.096, 4.124, 4.152, 4.18 , 4.208, 4.236, 4.264, 4.292, 4.32 , 4.348, 4.376, 4.404, 4.432, 4.46 , 4.488, 4.516, 4.544, 4.572, 4.6 , 4.628, 4.656, 4.684, 4.712, 4.74 , 4.768, 4.796, 4.824, 4.852, 4.88 , 4.908, 4.936, 4.964, 4.992, 5.02 , 5.048, 5.076, 5.104, 5.132, 5.16 , 5.188, 5.216, 5.244, 5.272, 5.3 , 5.328, 5.356, 5.384, 5.412, 5.44 , 5.468, 5.496, 5.524, 5.552, 5.58 , 5.608, 5.636, 5.664, 5.692, 5.72 , 5.748, 5.776, 5.804, 5.832, 5.86 , 5.888, 5.916, 5.944, 5.972, 6. , 6.028, 6.056, 6.084, 6.112, 6.14 , 6.168, 6.196, 6.224, 6.252, 6.28 , 6.308, 6.336, 6.364, 6.392, 6.42 , 6.448, 6.476, 6.504, 6.532, 6.56 , 6.588, 6.616, 6.644, 6.672, 6.7 , 6.728, 6.756, 6.784, 6.812, 6.84 , 6.868, 6.896, 6.924, 6.952, 6.98 , 7.008, 7.036, 7.064, 7.092, 7.12 , 7.148, 7.176, 7.204, 7.232, 7.26 , 7.288, 7.316, 7.344, 7.372, 7.4 , 7.428, 7.456, 7.484, 7.512, 7.54 , 7.568, 7.596, 7.624, 7.652, 7.68 , 7.708, 7.736, 7.764, 7.792, 7.82 , 7.848, 7.876, 7.904, 7.932, 7.96 , 7.988, 8.016, 8.044, 8.072, 8.1 , 8.128, 8.156, 8.184, 8.212, 8.24 , 8.268, 8.296, 8.324, 8.352, 8.38 , 8.408, 8.436, 8.464, 8.492, 8.52 , 8.548, 8.576, 8.604, 8.632, 8.66 , 8.688, 8.716, 8.744, 8.772, 8.8 , 8.828, 8.856, 8.884, 8.912, 8.94 , 8.968, 8.996, 9.024, 9.052, 9.08 , 9.108, 9.136, 9.164, 9.192, 9.22 , 9.248, 9.276, 9.304, 9.332, 9.36 , 9.388, 9.416, 9.444, 9.472, 9.5 ])
- (wavelength)float64𝟙0.206, 0.294, ..., nan, nanσ = 0.041, 0.053, ..., nan, nan
Values:
array([0.20606583, 0.2944515 , 0.2668158 , 0.24741376, 0.25958 , 0.26153507, 0.20935336, 0.19311085, 0.22279689, 0.22121681, 0.21579365, 0.21041809, 0.25755019, 0.23664326, 0.2712833 , 0.2965993 , 0.26083396, 0.20081185, 0.22184225, 0.19499865, 0.24042326, 0.23831378, 0.17726007, 0.24007785, 0.21214669, 0.2007215 , 0.23821212, 0.21950253, 0.22414521, 0.22740931, 0.239898 , 0.2062053 , 0.19817524, 0.19286924, 0.23283704, 0.19674343, 0.20275098, 0.22567763, 0.22986596, 0.22685141, 0.13523571, 0.16118647, 0.16071377, 0.17733342, 0.18219584, 0.28995007, 0.26772467, 0.18808852, 0.17112152, 0.19662742, 0.22447637, 0.24012482, 0.23178688, 0.19332748, 0.20747772, 0.24948179, 0.24670767, 0.19114659, 0.17772988, 0.18835225, 0.17464505, 0.18043201, 0.18546514, 0.19858774, 0.18472824, 0.23922206, 0.23585242, 0.19114568, 0.17956995, 0.19261109, 0.19873757, 0.19124756, 0.17718774, 0.18656745, 0.18385658, 0.20078546, 0.17624055, 0.19310344, 0.16276814, 0.16814312, 0.21495157, 0.1815441 , 0.16717576, 0.16675867, 0.15549182, 0.1536407 , 0.15042449, 0.15786886, 0.16256271, 0.16063645, 0.12733079, 0.16665954, 0.15521253, 0.14374998, 0.12852772, 0.13735643, 0.14798511, 0.11838767, 0.14415226, 0.10413252, 0.12286336, 0.10020518, 0.13801511, 0.13467133, 0.11976371, 0.13312148, 0.23681756, 0.33312826, 0.30774816, 0.31598602, 0.32790506, 0.25893835, 0.29485067, 0.28366428, 0.31160295, 0.29722834, 0.28540274, 0.32653728, 0.27961534, 0.28277563, 0.31081626, 0.33662039, 0.30503547, 0.29464245, 0.29762129, 0.28869478, 0.28855915, 0.2713608 , 0.31528965, 0.27700826, 0.29171888, 0.29503261, 0.30879049, 0.29881345, 0.29730408, 0.2609623 , 0.26845192, 0.27527503, 0.26219454, 0.28420303, 0.31664042, 0.28500055, 0.25428207, 0.25647223, 0.28100441, 0.27269992, 0.27775204, 0.31681621, 0.25613243, 0.24321132, 0.26104734, 0.30330042, 0.28769953, 0.268747 , 0.2495252 , 0.23872612, 0.28744429, 0.26321923, 0.30782821, 0.26289115, 0.25350435, 0.25264982, 0.25646106, 0.26752224, 0.23032663, 0.28920983, 0.25322182, 0.2416866 , 0.24034263, 0.22973101, 0.25126338, 0.24735726, 0.24814044, 0.26473432, 0.24912558, 0.25342644, 0.25304911, 0.27082889, 0.25974035, 0.24588537, 0.2616127 , 0.23248283, 0.21104377, 0.24038345, 0.23941452, 0.24690242, 0.24727856, 0.22525409, 0.23407253, 0.2342585 , 0.22542663, 0.24147349, 0.22851773, 0.25251838, 0.23498268, 0.23822337, 0.22853162, 0.22209972, 0.23429046, 0.2303793 , 0.22183052, 0.20702459, 0.21666083, 0.17292194, 0.21953968, 0.21773162, 0.21808337, 0.20723148, 0.22174483, 0.22771555, 0.21428222, 0.22689432, 0.21095777, 0.19931235, 0.21002884, 0.22338257, 0.22144632, 0.23267152, 0.21892891, 0.21183044, 0.22096842, 0.20571649, 0.21531391, 0.22682186, 0.20876636, 0.20401874, 0.20432734, 0.21673519, 0.20468401, 0.2078612 , 0.18818275, 0.18443707, 0.21079568, 0.19903931, 0.19818259, 0.1773508 , 0.20553103, 0.20193756, 0.20266248, 0.18855988, 0.18442355, 0.18876745, 0.19749294, 0.1939216 , 0.1993379 , 0.1900551 , 0.18910917, 0.16617635, nan, nan, 0.16247447, 0.20414137, 0.16923648, 0.18744141, 0.18973591, 0.18529801, 0.18843423, 0.19640482, 0.18044863, 0.19246188, 0.20107545, 0.17727981, 0.17849551, 0.18384203, 0.18830994, 0.16369439, 0.17959559, 0.16542227, 0.1623652 , 0.16994645, 0.18422343, 0.17491736, 0.16389896, 0.17836469, 0.18401206, 0.16182255, 0.17418692, 0.17566858, 0.16107084, 0.16557721, 0.17423476, 0.16794726, 0.17795251, 0.17638207, 0.15906307, 0.169131 , 0.17231668, 0.17000367, 0.16933696, 0.16425225, 0.15797136, 0.1646908 , 0.15761565, 0.16386056, 0.14586876, 0.16939056, nan, nan, nan, nan])
Variances (σ²):
array([1.70232857e-03, 2.81754808e-03, 2.31056864e-03, 2.20160752e-03, 1.73662771e-03, 2.06944143e-03, 1.16476280e-03, 9.72827851e-04, 1.18260126e-03, 1.15463344e-03, 1.02898692e-03, 1.02466279e-03, 1.45561530e-03, 1.13841656e-03, 1.34895364e-03, 1.51552440e-03, 1.09959302e-03, 7.10608776e-04, 7.64913047e-04, 6.49804441e-04, 8.78814153e-04, 7.88974490e-04, 4.69414403e-04, 7.61693544e-04, 6.37284915e-04, 5.34549455e-04, 6.53967000e-04, 5.47426379e-04, 5.57306210e-04, 5.76103820e-04, 6.39166331e-04, 4.98646056e-04, 4.41781396e-04, 4.28052421e-04, 5.73358031e-04, 4.32553473e-04, 4.93679255e-04, 5.94414488e-04, 7.13331730e-04, 6.95198447e-04, 2.85028566e-04, 4.13747385e-04, 4.50354095e-04, 6.29516003e-04, 6.67766896e-04, 1.43816490e-03, 1.25796335e-03, 7.31131149e-04, 8.44764433e-04, 1.22284981e-03, 1.33004453e-03, 1.05029489e-03, 1.05419428e-03, 7.00573015e-04, 7.00417583e-04, 8.74966748e-04, 7.27643912e-04, 5.13219965e-04, 4.64920607e-04, 4.61150503e-04, 4.11426991e-04, 4.15613180e-04, 4.77767091e-04, 4.33190859e-04, 3.63496764e-04, 5.72472166e-04, 5.26676957e-04, 4.03863820e-04, 3.28830649e-04, 3.44402944e-04, 3.78886847e-04, 3.68399957e-04, 3.19291887e-04, 3.41444024e-04, 3.19626444e-04, 3.77753341e-04, 3.15734436e-04, 3.54519799e-04, 2.36644668e-04, 2.62981733e-04, 3.94158012e-04, 3.03848378e-04, 2.51375288e-04, 2.51227713e-04, 2.31953361e-04, 2.19016780e-04, 2.07314226e-04, 2.16824269e-04, 2.43751457e-04, 2.32577184e-04, 1.69227443e-04, 2.39652705e-04, 2.13225283e-04, 1.99806608e-04, 1.62909832e-04, 1.94300200e-04, 2.00474448e-04, 1.44198501e-04, 1.93723249e-04, 1.23688965e-04, 1.61315076e-04, 1.44581763e-04, 2.70756616e-04, 1.72395837e-04, 1.30018685e-04, 1.79025452e-04, 3.59407107e-04, 5.29768404e-04, 4.48425872e-04, 4.29603888e-04, 4.53131724e-04, 3.08352692e-04, 3.63187442e-04, 3.59932875e-04, 4.05124465e-04, 3.56806458e-04, 3.23680700e-04, 4.05914622e-04, 2.99199060e-04, 3.16087286e-04, 3.53101156e-04, 4.11142475e-04, 3.67110820e-04, 3.14748141e-04, 3.18346179e-04, 3.07087058e-04, 3.08026127e-04, 2.65861907e-04, 3.47416541e-04, 2.90642073e-04, 3.08406680e-04, 2.98933575e-04, 3.21776914e-04, 3.15073108e-04, 3.15912330e-04, 2.47975109e-04, 2.65030275e-04, 2.50180010e-04, 2.42657374e-04, 2.63582632e-04, 3.30209502e-04, 2.87510675e-04, 2.18239008e-04, 2.25830746e-04, 2.83457492e-04, 2.45606701e-04, 2.88983728e-04, 3.48547044e-04, 2.43695734e-04, 2.45383825e-04, 2.58196541e-04, 3.47954427e-04, 4.08993580e-04, 4.54379050e-04, 4.81792755e-04, 4.04111809e-04, 3.76330521e-04, 2.90226581e-04, 2.92248695e-04, 2.12095440e-04, 1.95919681e-04, 1.88373675e-04, 1.96544924e-04, 2.09369073e-04, 1.66515462e-04, 2.33340841e-04, 1.80149946e-04, 1.78055704e-04, 1.64221942e-04, 1.58887417e-04, 1.67326795e-04, 1.79893062e-04, 1.67269788e-04, 2.01114517e-04, 1.72408250e-04, 1.78054107e-04, 1.75436439e-04, 2.00585555e-04, 1.71958306e-04, 1.53446859e-04, 2.00061988e-04, 1.50033122e-04, 1.24802432e-04, 1.62632809e-04, 1.58083387e-04, 1.59429650e-04, 1.68090670e-04, 1.37367711e-04, 1.49702607e-04, 1.50616269e-04, 1.39562116e-04, 1.67199753e-04, 1.53367887e-04, 1.62758238e-04, 1.63208460e-04, 1.62698517e-04, 1.52218078e-04, 1.46029690e-04, 1.64474863e-04, 1.78027489e-04, 2.03941400e-04, 2.50361579e-04, 7.89261411e-04, 1.56690971e-03, 2.65467087e-04, 1.73729780e-04, 1.58577935e-04, 1.24607998e-04, 1.17432525e-04, 1.41401458e-04, 1.21839143e-04, 1.32632832e-04, 1.08892330e-04, 9.79025151e-05, 1.22225328e-04, 1.19103988e-04, 1.23673044e-04, 1.29468908e-04, 1.13236695e-04, 1.10555218e-04, 1.16320799e-04, 9.91505791e-05, 1.16299275e-04, 1.28991997e-04, 1.08159051e-04, 1.02809014e-04, 1.02356646e-04, 1.20879757e-04, 9.71139087e-05, 1.01533343e-04, 9.40772409e-05, 9.02706103e-05, 1.06991798e-04, 9.47228079e-05, 8.80150941e-05, 8.58316683e-05, 9.88626108e-05, 1.03524920e-04, 1.03320253e-04, 8.85234727e-05, 9.35067257e-05, 8.45789731e-05, 9.58615606e-05, 9.74525440e-05, 1.07188716e-04, 1.17369740e-04, 1.83012889e-04, 2.48734532e-04, nan, nan, 3.46042359e-04, 2.26434151e-04, 1.34617185e-04, 1.15384879e-04, 1.08454675e-04, 9.75194688e-05, 1.03477356e-04, 1.20779942e-04, 8.08448111e-05, 9.84798774e-05, 1.05982767e-04, 8.28392233e-05, 8.60572856e-05, 8.41351048e-05, 9.39946419e-05, 7.02718600e-05, 8.31991732e-05, 6.28608407e-05, 6.82916401e-05, 7.73610455e-05, 8.54565680e-05, 7.65253854e-05, 6.49584807e-05, 7.41129623e-05, 7.51284209e-05, 6.35711687e-05, 6.65845947e-05, 7.63737590e-05, 7.30897736e-05, 6.81445220e-05, 7.27497295e-05, 6.63416558e-05, 7.28989155e-05, 7.09177885e-05, 6.08575922e-05, 6.16301942e-05, 7.49007415e-05, 7.48238919e-05, 6.88035057e-05, 6.95362813e-05, 6.33098684e-05, 6.60026114e-05, 7.77819410e-05, 1.04339799e-04, 1.20924345e-04, 2.23501698e-04, nan, nan, nan, nan])
[18]:
normalized.plot()
[18]:
Save the final result#
[19]:
from scippneutron.io import save_xye
to_disk = normalized.copy(deep=False)
to_disk.coords['wavelength'] = sc.midpoints(to_disk.coords['wavelength'])
save_xye('fe_bragg_edge.xye', to_disk)